Estimate survival function.
svykm.RdEstimates the survival function using a weighted Kaplan-Meier estimator.
svykm(formula, design,se=FALSE, ...)
# S3 method for svykm
plot(x,xlab="time",ylab="Proportion surviving",
ylim=c(0,1),ci=NULL,lty=1,...)
# S3 method for svykm
lines(x,xlab="time",type="s",ci=FALSE,lty=1,...)
# S3 method for svykmlist
plot(x, pars=NULL, ci=FALSE,...)
# S3 method for svykm
quantile(x, probs=c(0.75,0.5,0.25),ci=FALSE,level=0.95,...)
# S3 method for svykm
confint(object,parm,level=0.95,...)Arguments
- formula
Two-sided formula. The response variable should be a right-censored
Survobject- design
survey design object
- se
Compute standard errors? This is slow for moderate to large data sets
- ...
in
plotandlinesmethods, graphical parameters- x
a
svykmorsvykmlistobject- xlab,ylab,ylim,type
as for
plot- lty
Line type, see
par- ci
Plot (or return, for
quantile) the confidence interval- pars
A list of vectors of graphical parameters for the separate curves in a
svykmlistobject- object
A
svykmobject- parm
vector of times to report confidence intervals
- level
confidence level
- probs
survival probabilities for computing survival quantiles (note that these are the complement of the usual
quantileinput, so 0.9 means 90% surviving, not 90% dead)
Value
For svykm, an object of class svykm for a single curve or svykmlist
for multiple curves.
Details
When standard errors are computed, the survival curve is actually the Aalen (hazard-based) estimator rather than the Kaplan-Meier estimator.
The standard error computations use memory proportional to the sample size times the square of the number of events. This can be a lot.
In the case of equal-probability cluster sampling without replacement the computations are essentially the same as those of Williams (1995), and the same linearization strategy is used for other designs.
Confidence intervals are computed on the log(survival) scale,
following the default in survival package, which was based on
simulations by Link(1984).
Confidence intervals for quantiles use Woodruff's method: the interval is the intersection of the horizontal line at the specified quantile with the pointwise confidence band around the survival curve.
References
Link, C. L. (1984). Confidence intervals for the survival function using Cox's proportional hazards model with covariates. Biometrics 40, 601-610.
Williams RL (1995) "Product-Limit Survival Functions with Correlated Survival Times" Lifetime Data Analysis 1: 171--186
Woodruff RS (1952) Confidence intervals for medians and other position measures. JASA 57, 622-627.
See also
predict.svycoxph for survival curves from a Cox model
Examples
data(pbc, package="survival")
pbc$randomized <- with(pbc, !is.na(trt) & trt>0)
biasmodel<-glm(randomized~age*edema,data=pbc)
pbc$randprob<-fitted(biasmodel)
dpbc<-svydesign(id=~1, prob=~randprob, strata=~edema, data=subset(pbc,randomized))
s1<-svykm(Surv(time,status>0)~1, design=dpbc)
s2<-svykm(Surv(time,status>0)~I(bili>6), design=dpbc)
plot(s1)
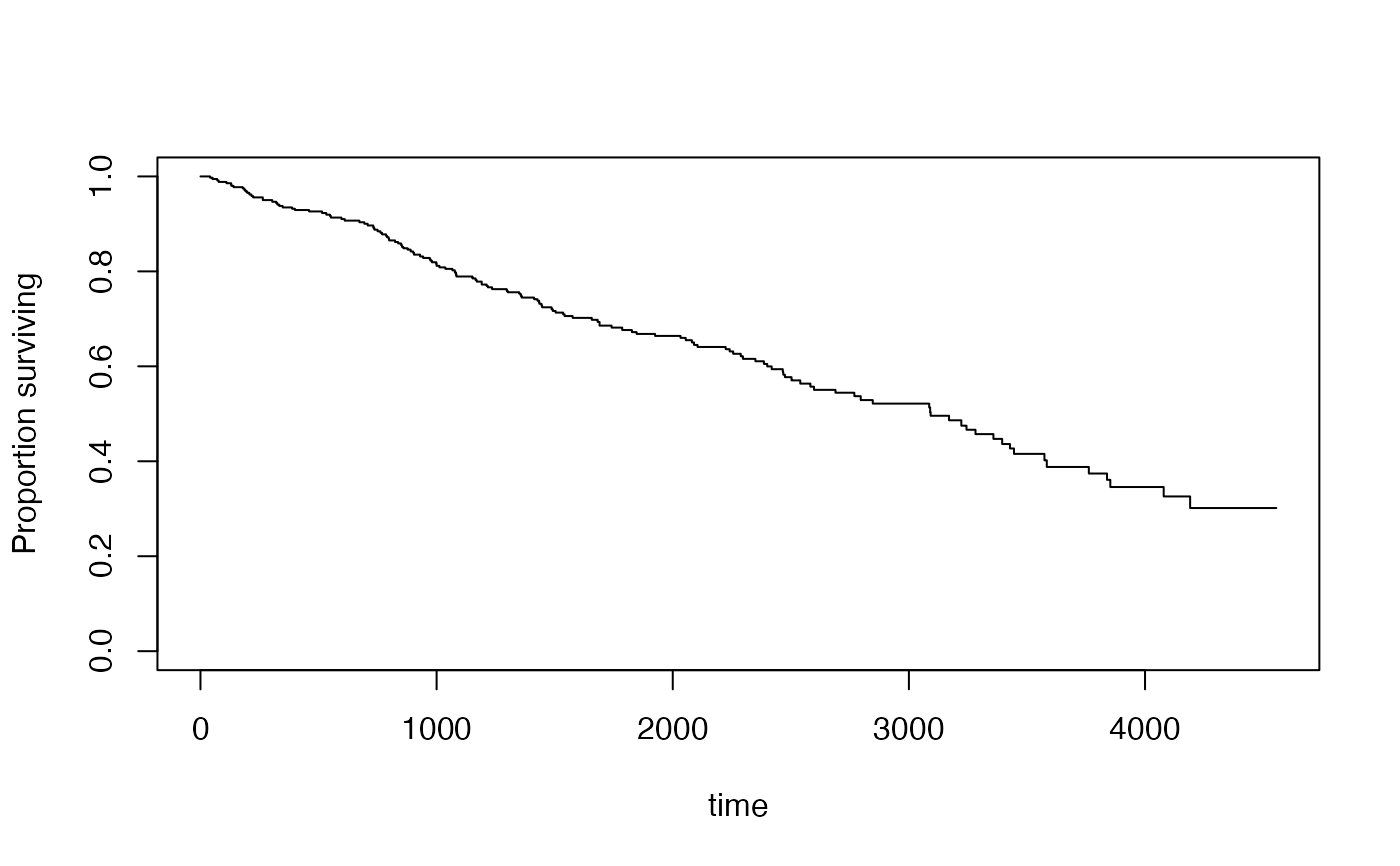 plot(s2)
plot(s2)
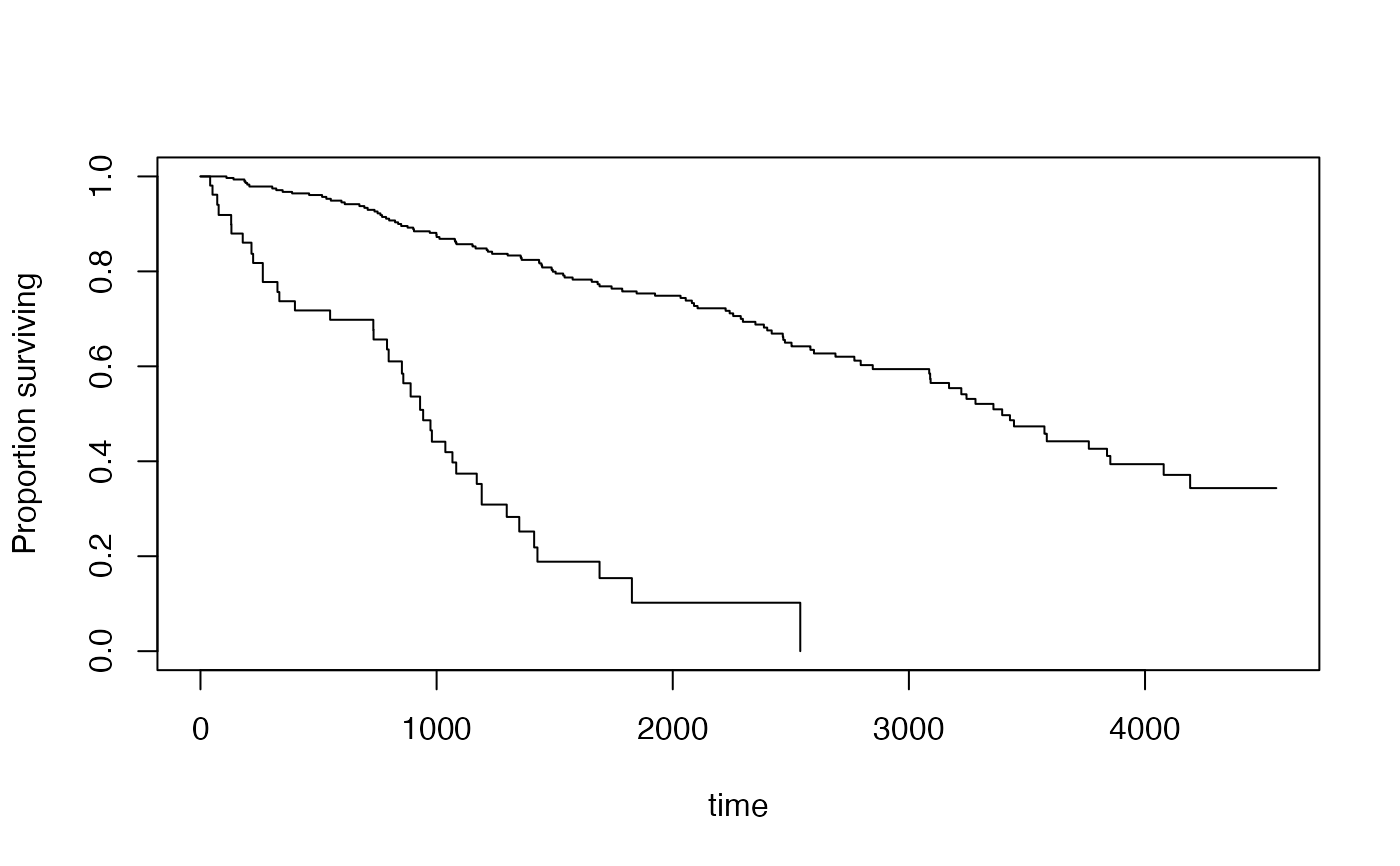 plot(s2, lwd=2, pars=list(lty=c(1,2),col=c("purple","forestgreen")))
plot(s2, lwd=2, pars=list(lty=c(1,2),col=c("purple","forestgreen")))
 quantile(s1, probs=c(0.9,0.75,0.5,0.25,0.1))
#> 0.9 0.75 0.5 0.25 0.1
#> 708 1356 3092 Inf Inf
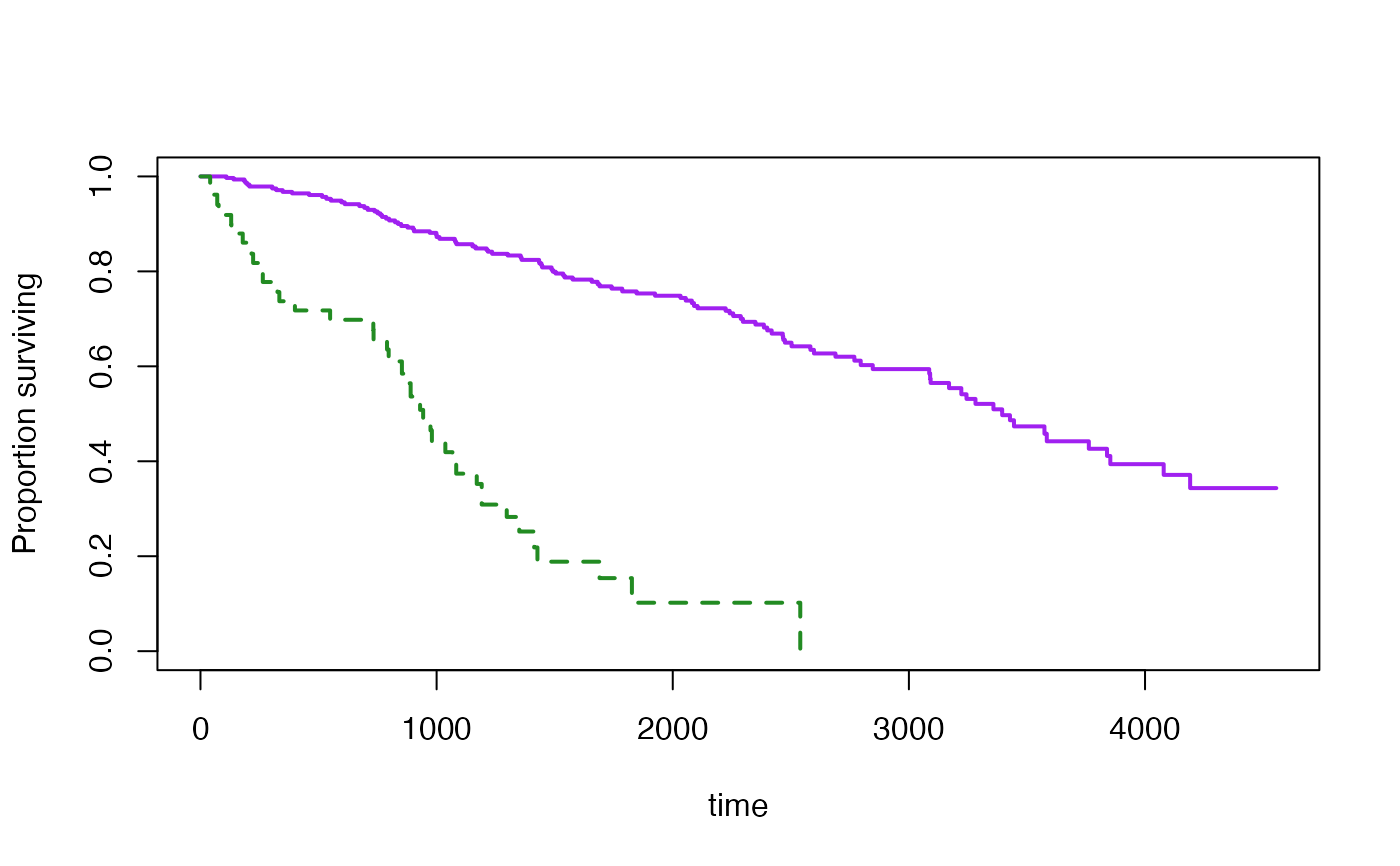
s3<-svykm(Surv(time,status>0)~I(bili>6), design=dpbc,se=TRUE)
plot(s3[[2]],col="purple")
quantile(s1, probs=c(0.9,0.75,0.5,0.25,0.1))
#> 0.9 0.75 0.5 0.25 0.1
#> 708 1356 3092 Inf Inf
s3<-svykm(Surv(time,status>0)~I(bili>6), design=dpbc,se=TRUE)
plot(s3[[2]],col="purple")
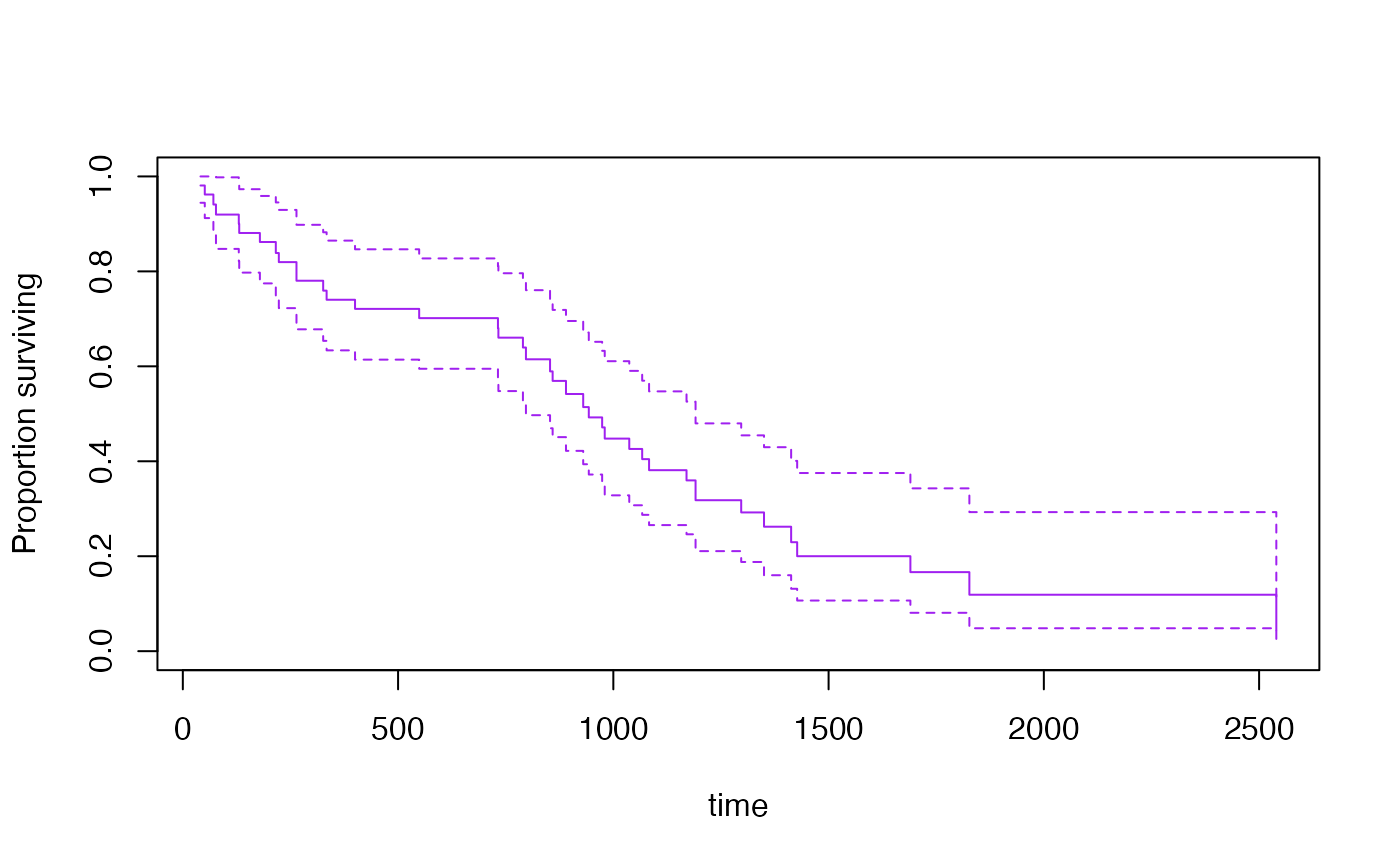 confint(s3[[2]], parm=365*(1:5))
#> 0.025 0.975
#> 365 0.63361572 0.8647973
#> 730 0.59486017 0.8272783
#> 1095 0.26557946 0.5472405
#> 1460 0.10663428 0.3752484
#> 1825 0.08082427 0.3429355
quantile(s3[[1]], ci=TRUE)
#> 0.75 0.5 0.25
#> 1925 3395 Inf
#> attr(,"ci")
#> 0.025 0.975
#> 0.75 1504 2386
#> 0.5 3092 4079
#> 0.25 Inf Inf
confint(s3[[2]], parm=365*(1:5))
#> 0.025 0.975
#> 365 0.63361572 0.8647973
#> 730 0.59486017 0.8272783
#> 1095 0.26557946 0.5472405
#> 1460 0.10663428 0.3752484
#> 1825 0.08082427 0.3429355
quantile(s3[[1]], ci=TRUE)
#> 0.75 0.5 0.25
#> 1925 3395 Inf
#> attr(,"ci")
#> 0.025 0.975
#> 0.75 1504 2386
#> 0.5 3092 4079
#> 0.25 Inf Inf